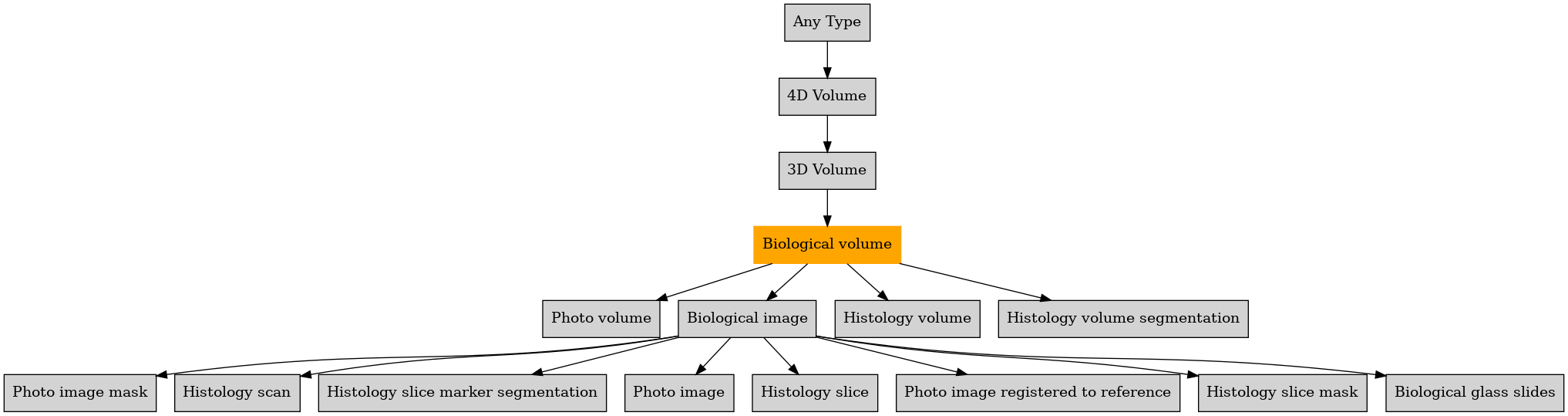

Defined in file : brainvisa/toolboxes/bioprocessing/types/biology.py
BrainVISA volume formats
{protocol}/{subject}/biology/anatomy/<subject>_nissl_volume
{protocol}/{subject}/biology/anatomy/<subject>_cytox_volume
{protocol}/{subject}/biology/anatomy/<subject>_ache_volume
{protocol}/{subject}/biology/functional/<subject>_autoradio_volume
{protocol}/{subject}/biology/{acquisition}/anatomy/<subject>_nissl_volume
{protocol}/{subject}/biology/{acquisition}/anatomy/<subject>_cytox_volume
{protocol}/{subject}/biology/{acquisition}/anatomy/<subject>_ache_volume
{protocol}/{subject}/biology/{acquisition}/functional/<subject>_autoradio_volume
protocol , subject , marker , category , acquisition , name_serie
study , center , subject , specie , photo_number , serie , scan_session , marker , extracted_level , scan_number , slice_number